Google’s AlphaGenome AI Cracks the DNA Code—Now Open-Source on GitHub
Google's latest AI breakthrough turns genetic sequences into something even Wall Street can understand—if they cared about science instead of memecoins.
### Decoding Life's Blueprint Just Got Easier
AlphaGenome slashes through DNA complexity like a hot wallet through weak security. No more squinting at nucleotide soup—this model translates ATCG into actionable insights faster than a crypto trader spotting a 10x opportunity.
### Open-Source Genomics for the Masses
GitHub's newest star repository puts genome analysis within reach of every biotech startup and basement researcher. Finally, an open-source project that might actually change lives—unlike most vaporware ICOs.
### The Fine Print
While hedge funds chase synthetic biology plays, real innovators are busy building the future. AlphaGenome won't cure cancer tomorrow, but it's the first AI that makes genetic data look less terrifying than a Bitcoin chart during a bear market.
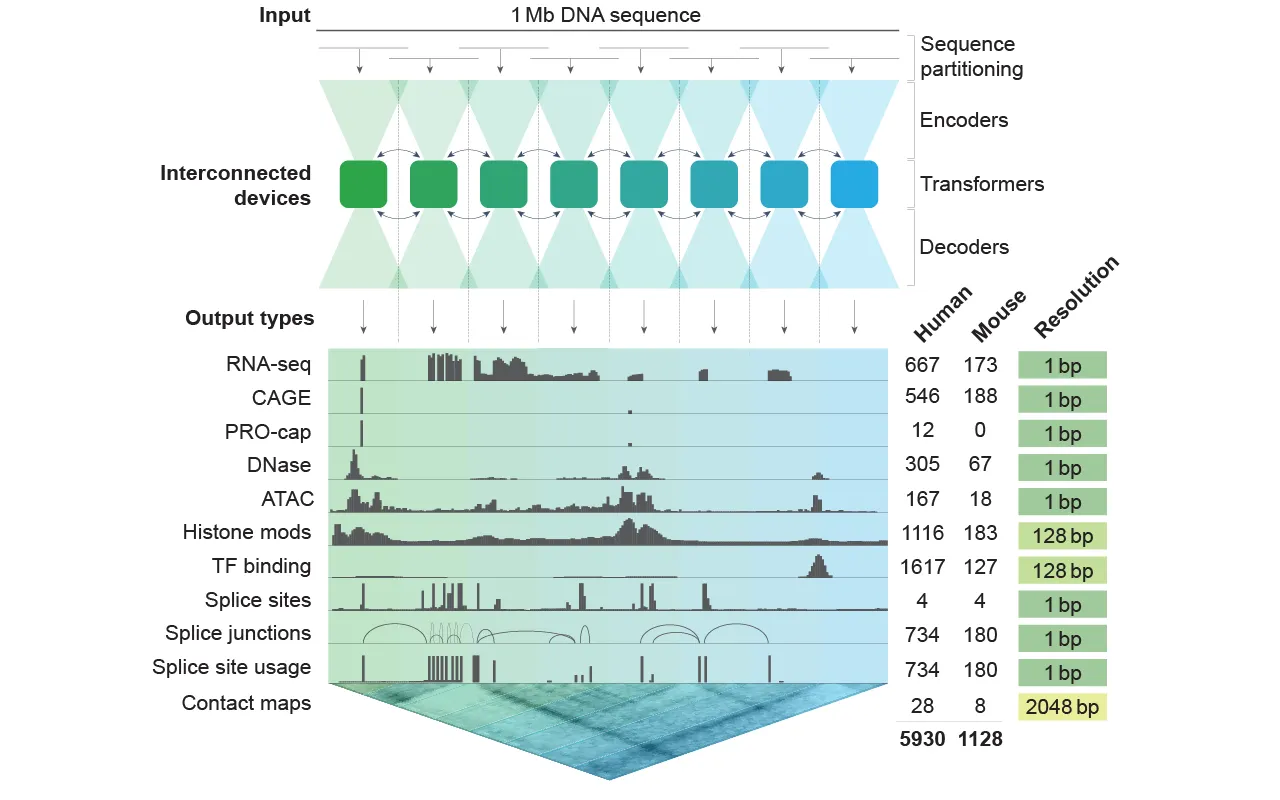
The architecture is notable, but still pretty familiar if you have been using Stable Diffusion or a normal open-source LLM locally: AlphaGenome uses a U-Net-inspired neural network, with about 450 million trainable parameters.
Yes, that is pretty low if you match it against even the weak and smaller language models that work with billions of parameters. However, considering that DNA only deals with 4 bases and only two pairs—basically the entire human genome is nothing but a combination of 3 billion pairs of A-T and C-G pairs of letters—it is a very specific model, designed to do one single thing extremely well.
The model has a sequence encoder that downsamples input from single-base resolution to coarser representations, then the transformer model layers long-range dependencies before the decoder reconstructs outputs back to the single-base level. This enables predictions at various resolutions, allowing for both fine-grained and broad regulatory analyses.
The model’s training relied on a wide array of publicly available datasets, including ENCODE, GTEx, 4D Nucleome, and FANTOM5—resources that collectively represent thousands of experimental profiles across human and mouse cell types.
And this process was also quite fast: using Google’s custom TPUs, DeepMind completed the pre-training and distillation process in just four hours, using half the computational budget required by its predecessor, Enformer.
AlphaGenome outperformed state-of-the-art models in 22 out of 24 sequence prediction tests and 24 out of 26 variant effect predictions, a rare clean sweep in benchmarks where incremental improvements are the norm. It does the job so well, in fact, that it can compare mutated and unmutated DNA, predicting the impact of genetic variants in seconds—a critical tool for researchers mapping disease origins.
This matters, because the non-coding genome contains many of the regulatory switches that control cell function and disease risk. Models like AlphaGenome are revealing how much of human biology is governed by these previously opaque regions.
AI’s influence on biology today is hard to ignore. Take Ankh, a protein language model developed by teams from the Technical University of Munich, Columbia University, and the startup Protinea. Ankh treats protein sequences like language, generating new proteins and predicting their behavior—similar to how AlphaGenome translates the regulatory “grammar” of DNA.
Another adjacent tech, Nvidia’s GenSLMs, demonstrates AI’s ability to forecast viral mutations and cluster genetic variants for pandemic research. Meanwhile, the use of AI to foster advances in chemical and gene-based anti-aging interventions highlights the intersection of genomics, machine learning, and medicine.
One of AlphaGenome’s most significant contributions is its accessibility. Rather than being restricted to commercial applications, the model is available via a public API for non-commercial research.
While it is not fully open sourced yet—meaning researchers can’t download and run or modify it locally—the API and accompanying resources allow scientists worldwide to generate predictions, adapt analyses for various species or cell types, and provide feedback to shape future releases. DeepMind has signaled plans for a broader open-source release down the line.
AlphaGenome’s ability to analyze non-coding variants—the area where most disease-linked mutations are found—could unlock new understanding of genetic disorders and rare diseases. Its high-speed variant scoring also supports personalized medicine, where treatments are tailored to an individual’s unique DNA profile.
For now, the non-coding genome is less of a black box, and AI’s role in genomics is set only to expand. AlphaGenome may not be the model to take us to Huxley’s "Brave New World," but it’s a clear sign of where things are headed: more data, better predictions, and a deeper understanding of how life works.

